Content available at: Indonesia (Indonesian) ไทย (Thai) Tiếng Việt (Vietnamese)
Newcastle Disease: Knowing the Virus Better to Make the Best Control Decisions. Part I
Newcastle disease (ND) is considered one of the most important infectious diseases of poultry because velogenic strains of the virus can cause outbreaks with high morbidity, mortality and restriction of international trade.
- This causes significant economic losses to the poultry industry. This is why the disease is included in the list of notifiable diseases to the World Organization for Animal Health (Miller and Koch, 2020).
- Newcastle disease is prevented by biosecurity and vaccination, and even though several types of effective live and inactivated vaccines are applied, ND continues to be a problem in many countries around the world.
THE CAUSAL AGENT
The International Committee on Viral Taxonomy database classifies the virus as family Paramyxoviridae, subfamily Avulavirinae, the latter distributed in three genera:
- Orthoavulavirus.
- Metaavulavirus.
- Paravulavirus.
Avian paramyxoviruses have been isolated from different avian species, and are classified into 21 serotypes by serological tests and phylogenetic analysis (WOAH, 2021).
- Newcastle disease is caused by virulent strains of Avian Paramyxovirus type 1 (APMV-1), species Avian Orthoavulavirus type -1 (OAV-1).
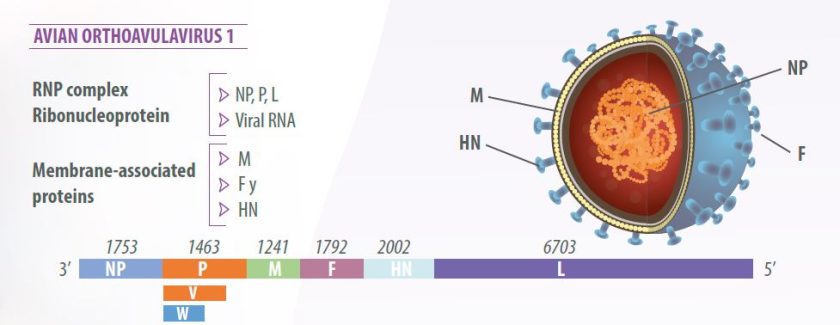
Newcastle disease virus (NDV) has a single-stranded, unsegmented, negative-sense RNA genome measuring 15,186 nucleotides (Alexander, 2003).
The virion has a lipid bilayer envelope derived from the plasma membrane of the host cell (Mast y Demeestere, 2009).
The virus genome is composed of six genes in 3’-NP-P-M-F-HN-L-5’ order, encoding seven viral proteins:
- Nucleoprotein (NP),
- Phosphoprotein (P),
- Matrix protein (M),
- Fusion protein (F),
- Hemagglutinin-neuraminidase (HN) and
- RNA polymerase, called the large polymerase (L).
RNA editing of the P protein produces an additional protein, the V protein, with anti-interferon activity, which allows the virus to counteract the innate host cell response (Miller and Koch, 2020) (Figure 1).
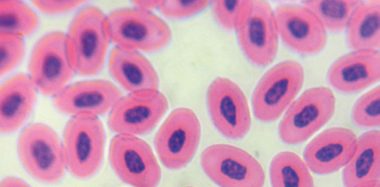
The main biological property of the virus is to agglutinate red blood cells of birds, amphibians and reptiles, due to the action of the HN protein on the sialic acid receptors on the surface of red blood cells.
Viral hemagglutination (HA) allows the determination of viral presence in viral cultures and allantoic fluid of chicken embryos (Miller and Koch, 2020), and the quantification of antibodies in bird serum by the hemagglutination inhibition test (HAI).
The oncolytic effect of some strains of NDV on human tumor cells and their use as a treatment for cancer in humans has been investigated for some time.
- This effect is associated with the anti-interferon type I activity of the V protein.
The selective replication that the virus has in tumor cells is due to defects of these cells in the activation of type I IFN signaling pathways and apoptotic pathways among others (Schirrmacher, 2017).
VIRAL REPLICATION
NDV replicates in the cytoplasm of the cell. Hemagglutinin (HN) recognizes the cellular receptor, activating the F protein to fuse the viral and cell membranes, allowing viral entry into the cytoplasm by endocytosis (Bergfeld, 2017; Miller and Koch, 2020).
- The ribonucleoprotein (RPN) complex contains the RNA genome wrapped with the nucleoprotein (NP) associated with the polymerase complex composed of the phosphoprotein (P) and the large protein (L);
- After entry of the viral nucleocapside into the cytoplasm, it dissociates from the M protein and is released to initiate the synthesis of the mRNA required for the translation of viral proteins.
- The P protein mediates the binding of the polymerase complex to the nucleocapside and the L protein performs the catalytic activities of the polymerase (Dortmans et al.,2011; Cox and Plemper, 2017).
- All genes code for a single major protein, however, the P gene mRNA results in the formation of two non-structural proteins, protein V and protein W (Vilela et al., 2022).
1. After the mRNA is translated into viral proteins, the negative polarity genome replicates, producing an anti-genomic RNA that serves as a template for the synthesis of a full-length genomic RNA (Miller and Koch, 2020; Dortmans et al., 2011).
2. HN proteins synthesized in the cell are transported to the cell membrane by insertion, followed by the alignment of the nucleocapside and viral RNA to the nearby modi ed regions of the cell membrane, which contain the viral glycoproteins (Miller and Koch, 2020);
- Matrix protein M, organizes the assembly of viral particles through interaction with N proteins of the RNP complex and membrane-integrated glycoprotein complexes (Cox and Plemper, 2017).
3. New viral particles bud out by budding from the cell surface, dragging the lipoprotein envelope of the cell (Miller and Koch, 2020; Cox and Plemper, 2017).
4. The neuraminidase of the HN protein allows the virus to be released from the cell by removing the cellular receptor (Bergfeld, 2017).
VIRAL PATHOGENICITY
According to the clinical signs they cause in infected birds, NDV strains are classified as follows:
- Velogenic viscerotropic strains (vvVEN) and mesogenic strains (both causing Newcastle disease) and,
- Lentogenic and enteric or apathogenic strains (used as vaccines).
The World Organization for Animal Health, WOAH, has determined that a strain of APMV- 1 is virulent when it has:
- An intracranial pathogenicity index (IPIC) equal to or greater than 0.7 (taking into consideration minimum and maximum values from 0 to 2) or,
- The F-protein cleavage site is multiple basic amino acids and has a phenylalanine at position 117 (WOAH, 2021).
F-glycoprotein is the key to viral virulence and pathogenesis, because cell entry depends on viral and cell membrane fusion after cleavage of the F0 protein into F1 and F2 by host proteases (Miller and Koch, 2020).
The difference between a virulent and a non-virulent virus is that: Viruses with a monobasic amino acid sequence at the F0 cleavage site are apathogenic because:
- This substrate is susceptible only to enzymes excreted in mucous membranes, such as trypsin, thus causing a localized infection.
Conversely, viruses with a dibasic sequence are pathogenic because they are:
- Susceptible to ubiquitous intracellular enzymes, which allow the virus to spread and cause systemic and fatal infection (Czeglédi et al., 2006; Miller and Koch, 2020).
INTRACRANIAL PATHOGENICITY INDEX
The intracranial pathogenicity index (IPIC) is the best method to determine viral pathogenicity because it not only determines the pathogenicity of the strains, but also quantifies in values.
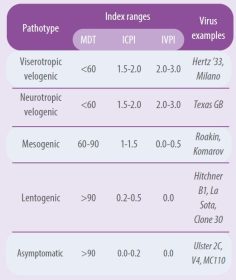
A virus is considered pathogenic when it has an IPIC equal to or greater than 0.7 (WOAH, 2021). There are great differences in pathogenicity among virus strains. Table 1 summarizes the pathogenicity index of the different pathotypes using different tests (Alexander, 1989 and 1998).
- Allantoic fluid con firmed positive for HA with a hemagglutinating titer greater than 1:16 is diluted 1/10 in saline solution and injected intracranially at a dose of 0.05 mL to ten 1-day-old SPF chicks.
- Birds are examined daily for 8 days, assigning them a daily score of: 0 if normal, 1 if sick and 2 if dead.
- Dead birds are recorded as 2 on each of the following days up to 8 days.
- IPIC is calculated as the average daily score per bird over the eight days; the most virulent viruses approach the maximum score of 2.0 and the lentogenic viruses approach 0.0.
All APMV-1 isolates are considered to belong to a single serotype, however, some antigenic variations have been demonstrated among different isolates by various tests and, even when there are no antigenic variations among strains, genetic analysis has become the main method of characterization and has replaced the use of mAbs for typing NDV isolates.
DETERMINATION OF VIRAL GENOTYPES
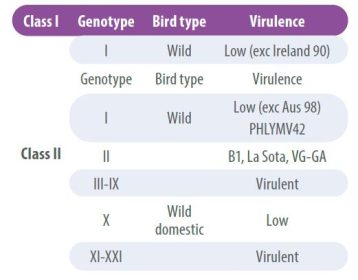
NDV viruses have low recombination rates, however, over time certain antigenic differences have been detected that have led to the classification of strains into lineages or genotypes. Accordingly, strains are grouped into two main classes, class I viruses and class II viruses;
- Class I viruses are of low pathogenicity and are found in wild birds.
- Class II, on the other hand, are classified into multiple genotypes and can be non-pathogenic or virulent (Table 2).
The characterization of the NDV genotype is achieved by sequencing the complete F gene, which also allows determining its virulence;
- To identify new genotypes a system based on a average evolutionary distance of 10% between genetic groups is used (Dimitrov KM et al 2016; Dimitrov KM et al, 2019).
The development of next-generation sequencing (NGS) has emerged as a tool for research and genetic characterization of pathogens.
- However, this method is ideal for rapidly mutating RNA viruses, where evolutionary aspects also need to be analyzed (Butt et al., 2019).
The current genotype classification includes three new genotypes (XIX, XX and XXI), making a total of 21 viral genotypes within class II viruses (Dimitrov et al. (2019).
RESISTANCE TO PHYSICAL AND CHEMICAL AGENTS
Being an enveloped virus, the viability of avian Paramyxoviruses is easily destroyed by physical and chemical agents such as:
- Heat,
- Ultraviolet light,
- Oxidation processes,
- pH and
- Treatment with most disinfectants.
However, publications on virus survival times outside the host show differences attributed to temperature, humidity and the environment in which the virus was analyzed (Kinde et al., 2004).











