Content available at: Español (Spanish) Português (Portuguese (Brazil))
Chickens are an ideal model organism for studies in phylogeny, embryology, medicine, and other various areas of scientific research.
Chicken protein is known for having a low degree of fat, a high degree of unsaturated fatty acids and low levels of sodium and cholesterol, which responds to current consumer demand.
Domestic chicken (Gallus gallus domesticus) is one of the main sources of highquality protein for humans
In order to obtain optimal patterns of high quality meat production, great advances have been made in nutrition and management in chickens.
However, a large part of these advances come from the high pressure of selection of animals in genetic improvement programs to obtain optimal rates of:
- Ready to sell carcass
- Efficiency
- Growth
The use of molecular techniques not only contributes to the improvement of selection, but also to the understanding of the evolutionary history of birds and of the genetic and epigenetic mechanisms involved in this evolutionary process and in the genetic diversification of this species.
This understanding is also important in a humanitarian context to improve the needs of animals and their breeding environments, as current models of large-scale production are questioned in relation to animal health and welfare.
Based on this, our research group in Brazil joined forces in collaboration with a research group in Sweden for the optimisation of molecular techniques to identify markers that will help us understand the genes that control performance and well-being characteristics in chickens.
Next Generation Sequencing (NGS)
In order to understand the molecular mechanisms governing these features of interest, especially in the last decade, high-performance sequencing techniques, also known as second-generation sequencing (NGS, Next Generation Sequencing), emerged.
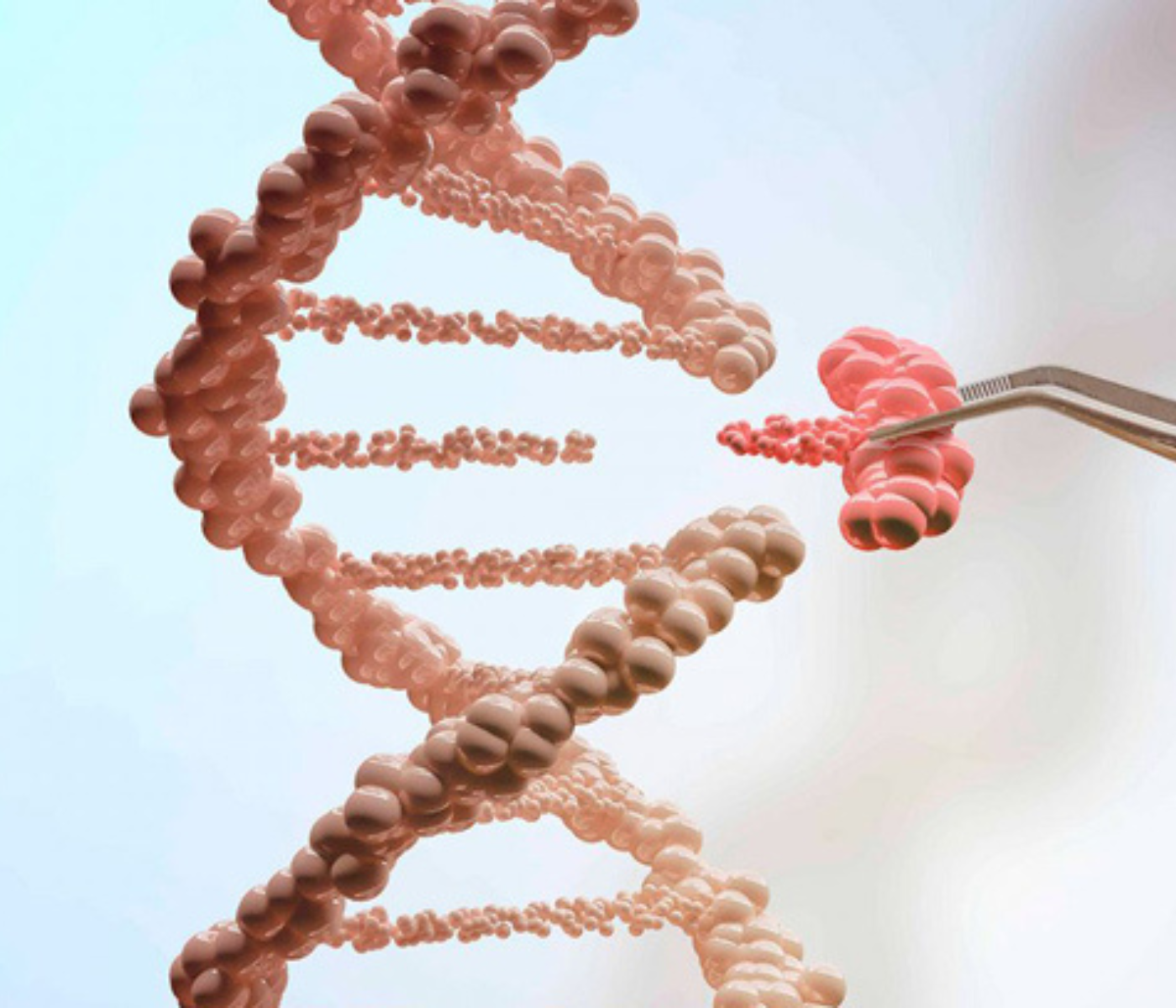
These methodologies provided a wealth of information that was used to identify both genetic mutation and epigenetic variations through molecular markers called single nucleotide polymorphisms (SNPs) and differential methylation regions in DNA (DMR).
Single nucleotide polymorphisms (SNPs) and differential methylation regions in DNA (DMR) may be responsible for functional changes in the chicken genome. They can be located in genome-neutral regions and are fundamental to many genetic activities and processes.
Second generation sequencing allowed the development of SNP panels to identify their associations with phenotypes of interest. However, these SNP panels have limited coverage in functionally important genomic regions in experimental populations.
Although NGS is powerful enough to detect informative polymorphisms, its high cost makes it impractical to use it in animal genetic improvement, in genomic wide association studies (GWAS) and in the detection of DMR by immunoprecipitation of methylated DNA (MeDIPS).
In this sense, taking into account the demand for the creation of a cost efficient method, we performed in silico simulations for the selection of a restriction enzyme (RE), which allowed the sequencing of a reduced fraction of the bird’s genome in an efficient, economical, and reproducible way for the discovery, characterisation and validation of SNPs and methylated regions.
This strategy made it possible to obtain fragments that present integral distribution and enrich regions of micro-chromosomes, which are underrepresented in commercially available genotyping panels.
The micro-chromosome regions are rich in genes with a high CpG* island content. The cytosine (C) of the CpG dinucleotides is susceptible to methylation in animals and can be used as an epigenetic marker.
Thus, based on an approach developed in Cornell, USA, we made a detailed step-by-step description of the complete optimisation of the reproducible protocol based on the reduced sequence of the chicken genome.
STUDY
From 462 animals genotyped using this protocol, it was possible to identify SNPs associated with performance characteristics in chickens.
Some of these SNPs had already been described as associated with the same characteristics in other populations, while others revealed new candidate regions.
Details of these genomic regions associated with animal performance were also published as a scientific paper.

The developed methodology also allowed a refinement in the technique of sequencing methylated DNA by immunoprecipitation (MeDIPseq), since our approach using ER showed a pattern of coverage in the genome that makes it unique compared to other approaches.
This profile includes not only an enrichment of different functional regions, but also a high focus on micro-chromosome regions that are CpG-rich regions and have higher gene density than macro-chromosomes.
This led us to consider the use of this methodology for the development of other studies involving access to methylation profiles of individuals.
Traditionally, the analysis of differentiated methylated regions (DMR) analyses the entire genome and has a high cost. Our discovery made it possible to reduce the price of the technique by using an ER derived from GBS optimisation*.
CpG enriched by restriction enzyme digestion can be used as sentinels for prolonged stress.
We use erythrocyte DNA from birds, since they are cells that are easily accessible to the field and easily isolated in blood tissue, under different rearing conditions (cage vs. open aviary).
We identified hundreds of differentiated methylated regions.
We can access these DMRs in the repository (EMBL-EBI) of the European Nucleotide Archive (ENA) under the access number PRJEB21356
CONCLUSION
In summary, our work resulted in the discovery of genomic regions associated with performance in chickens and in the measurement of stress in animals by quantifying differentiated methylated regions.
This methodology brings us very close to the development of a definitive tool for the diagnosis of stress in animals subjected to different production conditions. It also brings us closer to the direct application of these molecular tools in the production environment.
This work had the support of CAPES (Coordination for the Improvement of Higher Level Personnel – Brazil) and currently its continuity is supported by FAPESP (Foundation for Amparo to Research of the State of So Paulo)
References will be provided upon request
PDF